pacman::p_load(sf, spdep, tmap, tidyverse)In-Class Exercise 1: Spatial Weights and Applications
1 Overview
In this hands-on exercise, I will demonstrate computing spatial weights using R. The following steps are included in this exercise:
importing geospatial data using appropriate function(s) from sf package,
importing csv file using appropriate function from readr package,
performing relational join using appropriate join function from dplyr package,
computing spatial weights using appropriate functions from spdep package, and
calculating spatially lagged variables using appropriate functions from spdep package.
2 The Study Area and Data
The two data sets used in this hands-on exercise are:
Hunan county boundary layer. This is a geospatial data set in ESRI shapefile format.
Hunan_2012.csv: This csv file contains selected Hunan’s local development indicators in 2012.
2.1 Getting Started
3 Getting the Data into R Environment
A geospatial data and its associated attribute table will be loaded into the R environment. The geospatial data is in ESRI shapefile format and the attribute table is in csv fomat.
3.1 Import shapefile into r environment
The following code chunk imports the Hunan shapefile into R. The imported shapefile is a simple features object of sf package.
hunan <- st_read(dsn = "data/geospatial",
layer = "Hunan")Reading layer `Hunan' from data source
`C:\lohsiying\ISSS624\in_class_ex\ex1\data\geospatial' using driver `ESRI Shapefile'
Simple feature collection with 88 features and 7 fields
Geometry type: POLYGON
Dimension: XY
Bounding box: xmin: 108.7831 ymin: 24.6342 xmax: 114.2544 ymax: 30.12812
Geodetic CRS: WGS 843.2 Import csv file into R environment
In the following code chunk, imports a csv file using read_csv() of readr package to give an output in R dataframe class.
hunan2012 <- read_csv("data/aspatial/Hunan_2012.csv")Rows: 88 Columns: 29
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (2): County, City
dbl (27): avg_wage, deposite, FAI, Gov_Rev, Gov_Exp, GDP, GDPPC, GIO, Loan, ...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.3.3 Performing relational join
The code chunk updates the attribute table of hunan’s SpatialPolygonsDataFrame with the attribute fields of hunan2012 dataframe. This is performed by using left_join() from dplyr package.
hunan <- left_join(hunan,hunan2012)Joining, by = "County"4 Visualizing Regional Development Indicator
In here, a basemap and a choropleth map showing the distribution of GDPPC 2012 is prepared by using qtm() from tmap package.
basemap <- tm_shape(hunan) +
tm_polygons() +
tm_text("NAME_3", size=0.5)
gdppc <- qtm(hunan, "GDPPC")
tmap_arrange(basemap, gdppc, asp=1, ncol=2)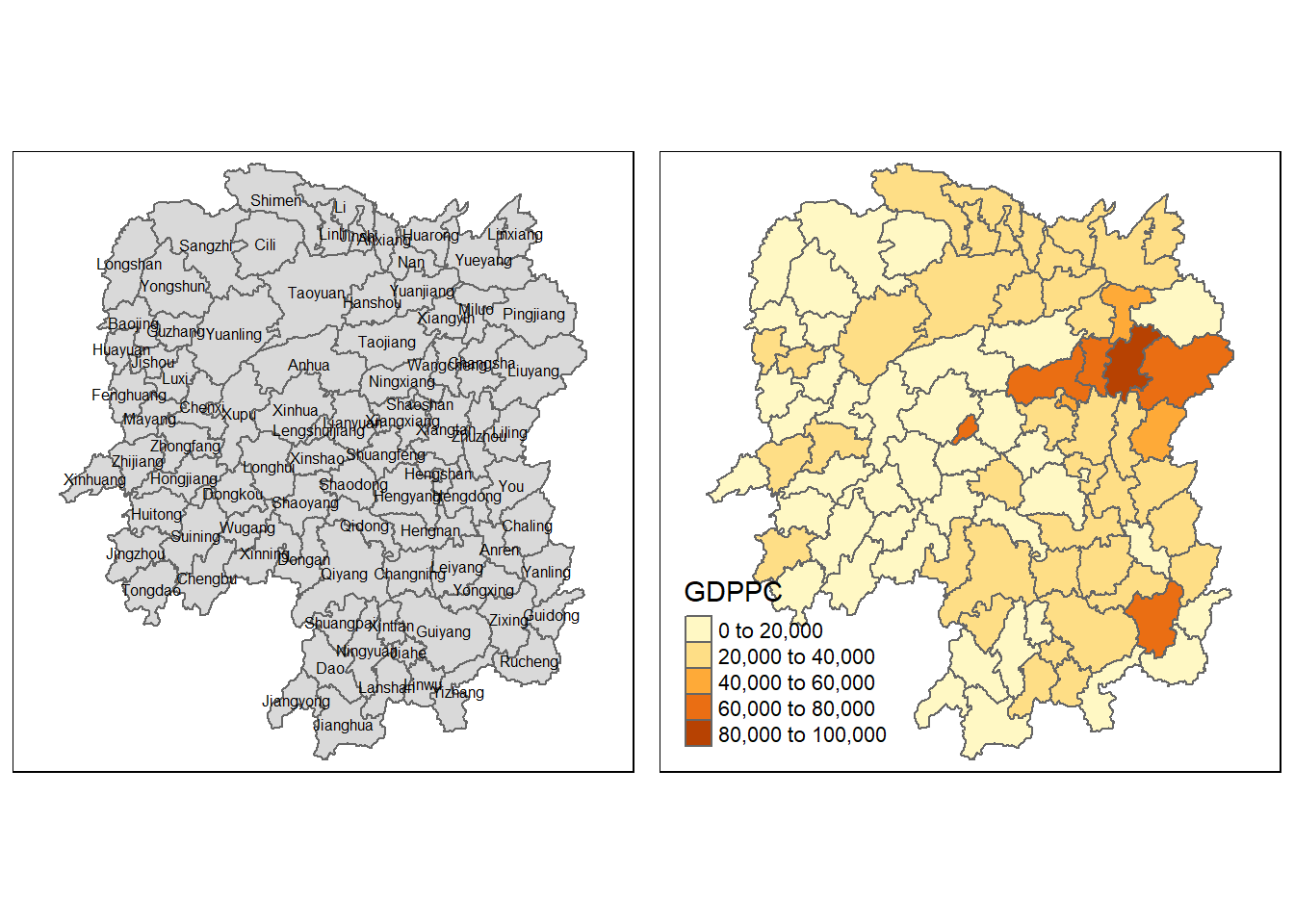
5 Computing Contiguity Spatial Weights
In this section, poly2nb()from spdep package is used to compute the contiguity weight matrices for the study area. By default, this function builds a list of neighbours based on regions with contiguous boundaries, using the Queen criteria. This based on the argument Queen set to True by default. Setting to False will enable Rook criteria.
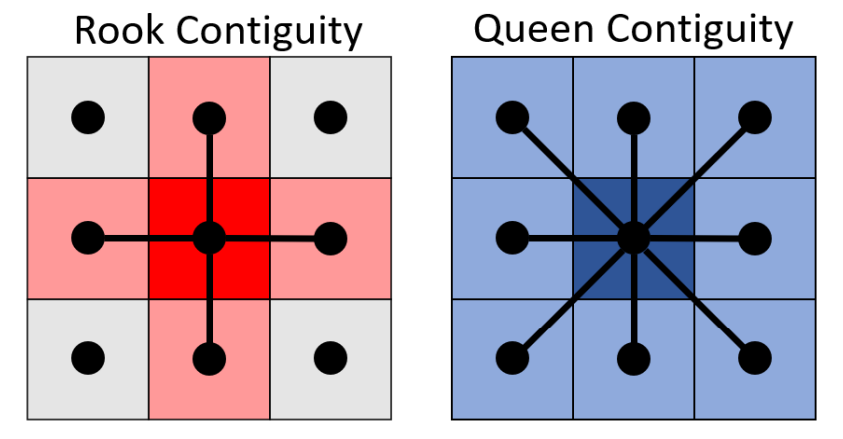
(Reference [1])
5.1 Computing (QUEEN) contiguity based neighbours
The code chunk below is used to compute the Queen contiguity weight matrix.
wm_q <- poly2nb(hunan, queen=TRUE)
summary(wm_q)Neighbour list object:
Number of regions: 88
Number of nonzero links: 448
Percentage nonzero weights: 5.785124
Average number of links: 5.090909
Link number distribution:
1 2 3 4 5 6 7 8 9 11
2 2 12 16 24 14 11 4 2 1
2 least connected regions:
30 65 with 1 link
1 most connected region:
85 with 11 linksThe summary report above shows that there are 88 area units in Hunan. The most connected area unit has 11 neighbours. There are two area units with only one neighbour.
For each polygon in our polygon object, wm_q lists all neighboring polygons. For example, in the following code chunk we can see the neighbors for the first polygon:
wm_q[[1]][1] 2 3 4 57 85Polygon 1 has 5 neighbors. The numbers represent the polygon IDs stored in hunan SpatialPolygonsDataFrame class.
We can retrive the county name of Polygon ID=1 by using the code chunk below:
hunan$County[1][1] "Anxiang"The output reveals that Polygon ID=1 is Anxiang county.
To reveal the county names of the five neighboring polygons, the following code chunk can be used:
hunan$NAME_3[c(2,3,4,57,85)][1] "Hanshou" "Jinshi" "Li" "Nan" "Taoyuan"We can retrieve the GDPPC of these five counties by using the code chunk below.
nb1 <- wm_q[[1]]
nb1 <- hunan$GDPPC[nb1]
nb1[1] 20981 34592 24473 21311 22879The printed output shows that the GDPPC of the five nearest neighbours based on Queen’s method are 20981, 34592, 24473, 21311 and 22879 respectively.
To display the complete weight matrix, str() can be used.
str(wm_q)List of 88
$ : int [1:5] 2 3 4 57 85
$ : int [1:5] 1 57 58 78 85
$ : int [1:4] 1 4 5 85
$ : int [1:4] 1 3 5 6
$ : int [1:4] 3 4 6 85
$ : int [1:5] 4 5 69 75 85
$ : int [1:4] 67 71 74 84
$ : int [1:7] 9 46 47 56 78 80 86
$ : int [1:6] 8 66 68 78 84 86
$ : int [1:8] 16 17 19 20 22 70 72 73
$ : int [1:3] 14 17 72
$ : int [1:5] 13 60 61 63 83
$ : int [1:4] 12 15 60 83
$ : int [1:3] 11 15 17
$ : int [1:4] 13 14 17 83
$ : int [1:5] 10 17 22 72 83
$ : int [1:7] 10 11 14 15 16 72 83
$ : int [1:5] 20 22 23 77 83
$ : int [1:6] 10 20 21 73 74 86
$ : int [1:7] 10 18 19 21 22 23 82
$ : int [1:5] 19 20 35 82 86
$ : int [1:5] 10 16 18 20 83
$ : int [1:7] 18 20 38 41 77 79 82
$ : int [1:5] 25 28 31 32 54
$ : int [1:5] 24 28 31 33 81
$ : int [1:4] 27 33 42 81
$ : int [1:3] 26 29 42
$ : int [1:5] 24 25 33 49 54
$ : int [1:3] 27 37 42
$ : int 33
$ : int [1:8] 24 25 32 36 39 40 56 81
$ : int [1:8] 24 31 50 54 55 56 75 85
$ : int [1:5] 25 26 28 30 81
$ : int [1:3] 36 45 80
$ : int [1:6] 21 41 47 80 82 86
$ : int [1:6] 31 34 40 45 56 80
$ : int [1:4] 29 42 43 44
$ : int [1:4] 23 44 77 79
$ : int [1:5] 31 40 42 43 81
$ : int [1:6] 31 36 39 43 45 79
$ : int [1:6] 23 35 45 79 80 82
$ : int [1:7] 26 27 29 37 39 43 81
$ : int [1:6] 37 39 40 42 44 79
$ : int [1:4] 37 38 43 79
$ : int [1:6] 34 36 40 41 79 80
$ : int [1:3] 8 47 86
$ : int [1:5] 8 35 46 80 86
$ : int [1:5] 50 51 52 53 55
$ : int [1:4] 28 51 52 54
$ : int [1:5] 32 48 52 54 55
$ : int [1:3] 48 49 52
$ : int [1:5] 48 49 50 51 54
$ : int [1:3] 48 55 75
$ : int [1:6] 24 28 32 49 50 52
$ : int [1:5] 32 48 50 53 75
$ : int [1:7] 8 31 32 36 78 80 85
$ : int [1:6] 1 2 58 64 76 85
$ : int [1:5] 2 57 68 76 78
$ : int [1:4] 60 61 87 88
$ : int [1:4] 12 13 59 61
$ : int [1:7] 12 59 60 62 63 77 87
$ : int [1:3] 61 77 87
$ : int [1:4] 12 61 77 83
$ : int [1:2] 57 76
$ : int 76
$ : int [1:5] 9 67 68 76 84
$ : int [1:4] 7 66 76 84
$ : int [1:5] 9 58 66 76 78
$ : int [1:3] 6 75 85
$ : int [1:3] 10 72 73
$ : int [1:3] 7 73 74
$ : int [1:5] 10 11 16 17 70
$ : int [1:5] 10 19 70 71 74
$ : int [1:6] 7 19 71 73 84 86
$ : int [1:6] 6 32 53 55 69 85
$ : int [1:7] 57 58 64 65 66 67 68
$ : int [1:7] 18 23 38 61 62 63 83
$ : int [1:7] 2 8 9 56 58 68 85
$ : int [1:7] 23 38 40 41 43 44 45
$ : int [1:8] 8 34 35 36 41 45 47 56
$ : int [1:6] 25 26 31 33 39 42
$ : int [1:5] 20 21 23 35 41
$ : int [1:9] 12 13 15 16 17 18 22 63 77
$ : int [1:6] 7 9 66 67 74 86
$ : int [1:11] 1 2 3 5 6 32 56 57 69 75 ...
$ : int [1:9] 8 9 19 21 35 46 47 74 84
$ : int [1:4] 59 61 62 88
$ : int [1:2] 59 87
- attr(*, "class")= chr "nb"
- attr(*, "region.id")= chr [1:88] "1" "2" "3" "4" ...
- attr(*, "call")= language poly2nb(pl = hunan, queen = TRUE)
- attr(*, "type")= chr "queen"
- attr(*, "sym")= logi TRUE5.2 Creating (ROOK) contiguity based neighbours
The code chunk below is used to compute Rook contiguity weight matrix.
wm_r <- poly2nb(hunan, queen=FALSE)
summary(wm_r)Neighbour list object:
Number of regions: 88
Number of nonzero links: 440
Percentage nonzero weights: 5.681818
Average number of links: 5
Link number distribution:
1 2 3 4 5 6 7 8 9 10
2 2 12 20 21 14 11 3 2 1
2 least connected regions:
30 65 with 1 link
1 most connected region:
85 with 10 linksThe summary report above shows that there are 88 area units in Hunan. Using the Rook’s method, the most connect area unit has 10 neighbours. There are two area units with only one neighbours.
5.3 Visualising contiguity weights
A connectivity graph takes a point and displays a line to each neighboring point. Now the data is in polygon format, so we will need to get points in order to make our connectivity graphs. The most typical method for this will be to use polygon centroids by specifying their Latitude and Longitude. We will calculate these in the sf package before moving onto the graphs.
We need the Latitude and Longitude coordinates in a separate data frame for this. To do this we will use a mapping function to apply a given function to each element of a vector and return a vector of the same length. Our input vector will be the geometry column of us.bound. Our function will be st_centroid. We will be using map_dbl variation of map from the purrr package.
To get our longitude values we map the st_centroid function over the geometry column of us.bound and access the longitude value through double bracket notation [[]] and 1. This allows us to get only the longitude, which is the first value in each centroid.
longitude <- map_dbl(hunan$geometry, ~st_centroid(.x)[[1]])We do the same for latitude with one key difference. We access the second value of each centroid with [[2]].
latitude <- map_dbl(hunan$geometry, ~st_centroid(.x)[[2]])Now that we have latitude and longitude, we use cbind to put longitude and latitude into the same object.
coords <- cbind(longitude, latitude)5.3.1 Plotting Queen contiguity based neighbours map
plot(hunan$geometry, border="lightgrey")
plot(wm_q, coords, pch = 19, cex = 0.6, add = TRUE, col = "red")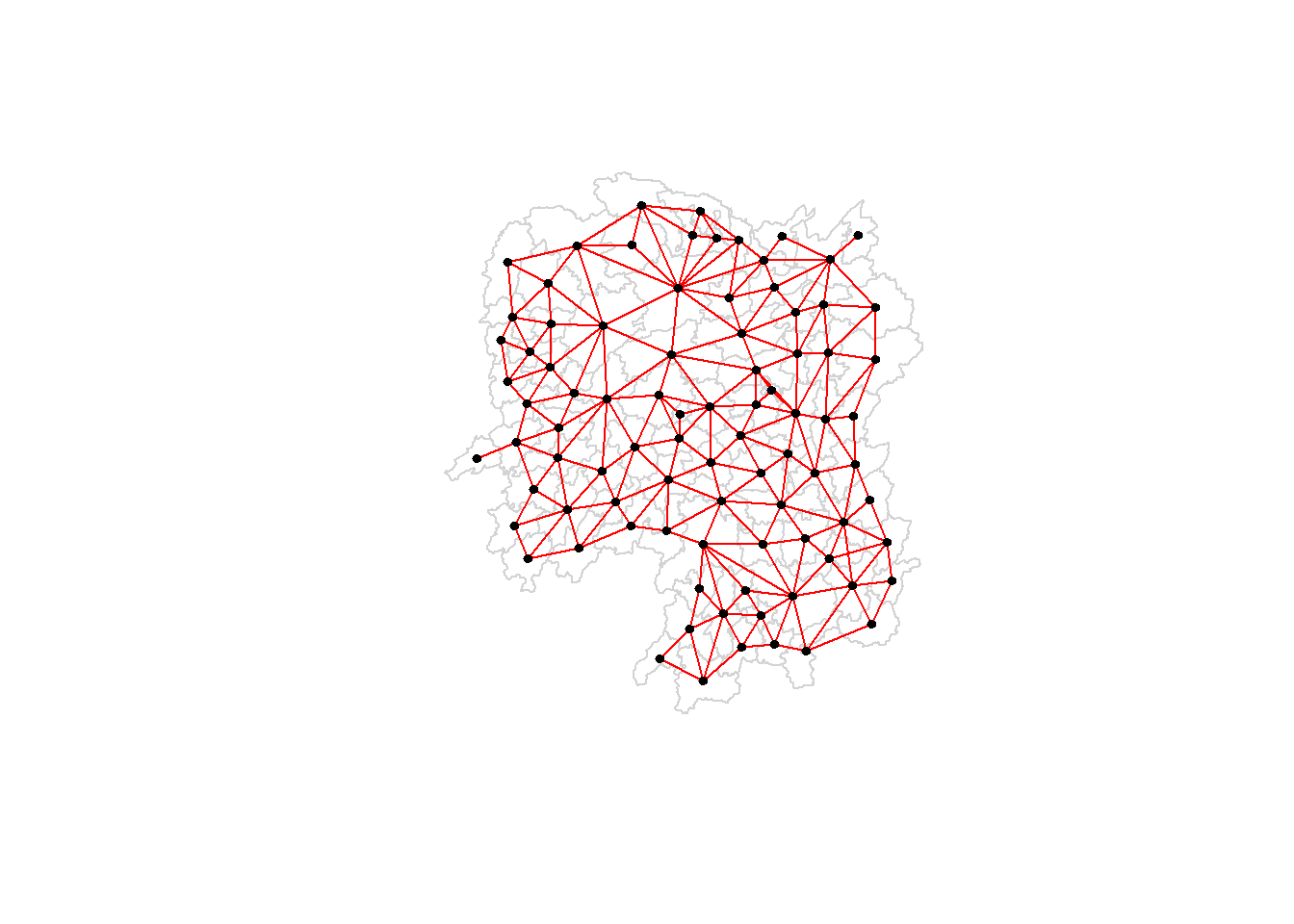
5.3.2 Plotting Rook contiguity based neighbours map
plot(hunan$geometry, border="lightgrey")
plot(wm_r, coords, pch = 19, cex = 0.6, add = TRUE, col = "red")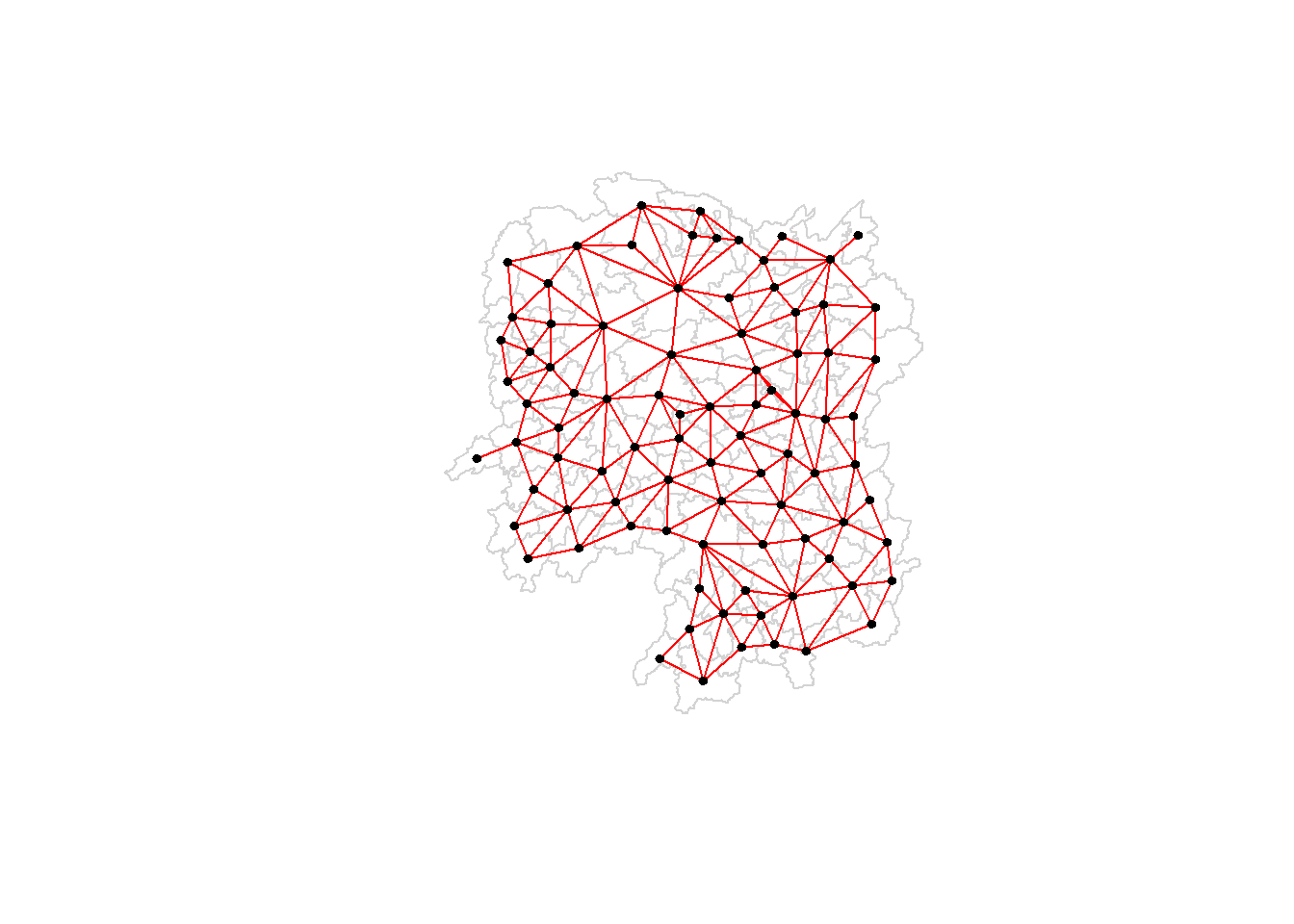
5.3.3 Plotting both Queen and Rook contiguity based neighbours maps
par(mfrow=c(1,2), mar = c(0, 0, 2, 3) + 0.1)
plot(hunan$geometry, border="lightgrey", main = 'Queen Contiguity')
plot(wm_q, coords, pch = 19, cex = 0.6, add = TRUE, col= "red")
plot(hunan$geometry, border="lightgrey", main = 'Rook Contiguity')
plot(wm_r, coords, pch = 19, cex = 0.6, add = TRUE, col = "red")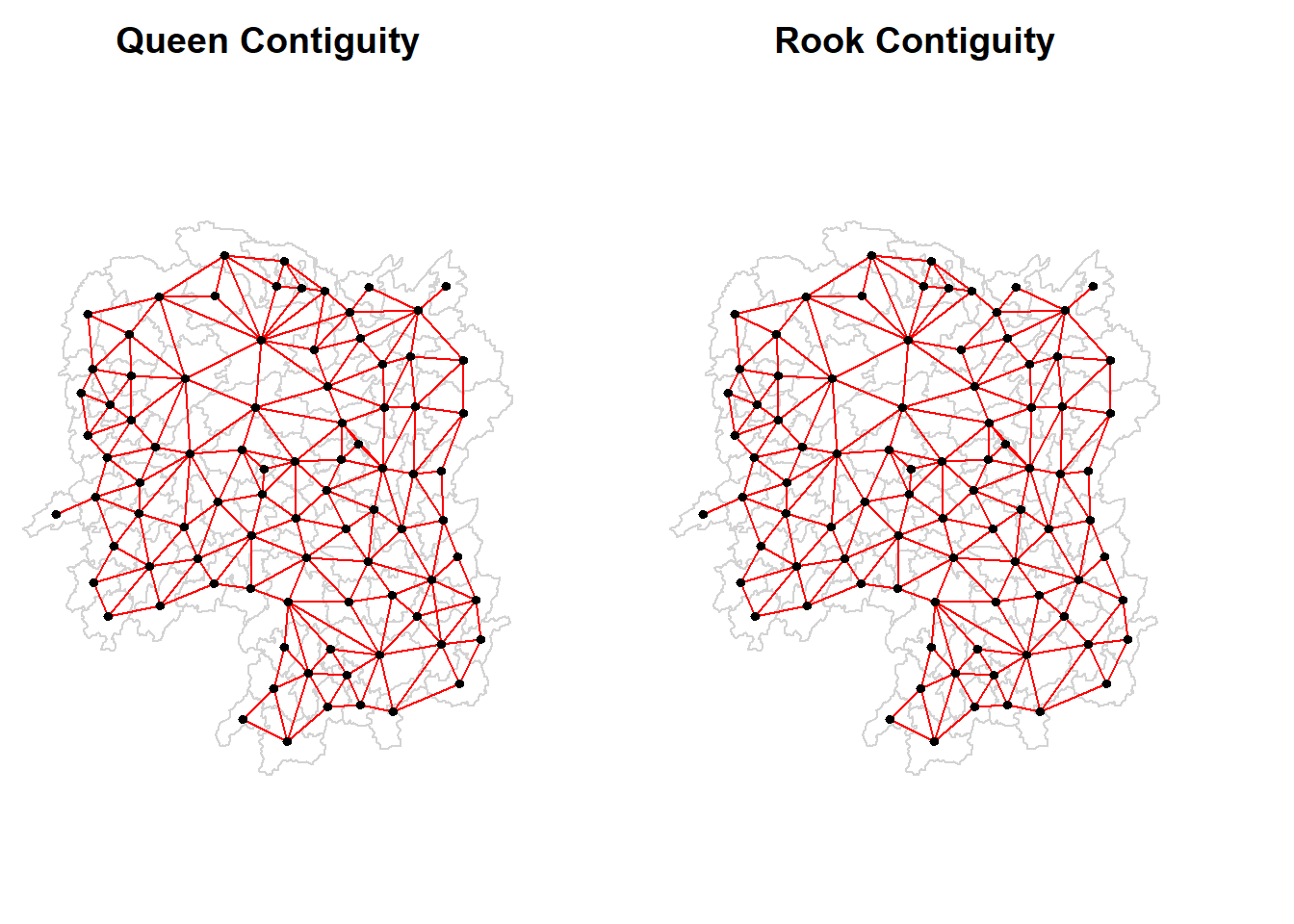
6 Computing distance based neighbours
In this section, distance-based weight matrices will be derived. This is performed by using dnearneigh() from spdep package.
The function identifies neighbours of region points by Euclidean distance in the metric of the points between lower (greater than or equal to) and upper (less than or equal to) bounds. If unprojected coordinates are used and either specified in the coordinates object x or with x as a two column matrix and longlat=TRUE, great circle distances in km will be calculated assuming the WGS84 reference ellipsoid.
6.1 Determine the cut-off distance
We will first determine the upper limit for distance band by using the steps below:
Return a matrix with the indices of points belonging to the set of the k nearest neighbours of each other by using knearneigh() of spdep.
Convert the knn object returned by knearneigh() into a neighbours list of class nb with a list of integer vectors containing neighbour region ids by using knn2nb().
Return the length of neighbour relationship edges by using nbdists() of spdep. The function returns in the units of the coordinates if the coordinates are projected, in km otherwise.
Remove the list structure of the returned object by using unlist().
#coords <- coordinates(hunan)
k1 <- knn2nb(knearneigh(coords))
k1dists <- unlist(nbdists(k1, coords, longlat = TRUE))
summary(k1dists) Min. 1st Qu. Median Mean 3rd Qu. Max.
24.79 32.57 38.01 39.07 44.52 61.79 The summary report shows that the largest first nearest neighbour distance is 61.79 km, so using this as the upper bound gives certainty that all units will have at least one neighbour.
6.2 Computing fixed distance weight matrix
Next, we will compute the distance weight matrix by using dnearneigh() as shown in the code chunk below.
wm_d62 <- dnearneigh(coords, 0, 62, longlat = TRUE)
wm_d62Neighbour list object:
Number of regions: 88
Number of nonzero links: 324
Percentage nonzero weights: 4.183884
Average number of links: 3.681818 The output above shows that on average, each region has 3.68 neighbours.
Next, we will use str() to display the content of wm_d62 weight matrix.
str(wm_d62)List of 88
$ : int [1:5] 3 4 5 57 64
$ : int [1:4] 57 58 78 85
$ : int [1:4] 1 4 5 57
$ : int [1:3] 1 3 5
$ : int [1:4] 1 3 4 85
$ : int 69
$ : int [1:2] 67 84
$ : int [1:4] 9 46 47 78
$ : int [1:4] 8 46 68 84
$ : int [1:4] 16 22 70 72
$ : int [1:3] 14 17 72
$ : int [1:5] 13 60 61 63 83
$ : int [1:4] 12 15 60 83
$ : int [1:2] 11 17
$ : int 13
$ : int [1:4] 10 17 22 83
$ : int [1:3] 11 14 16
$ : int [1:3] 20 22 63
$ : int [1:5] 20 21 73 74 82
$ : int [1:5] 18 19 21 22 82
$ : int [1:6] 19 20 35 74 82 86
$ : int [1:4] 10 16 18 20
$ : int [1:3] 41 77 82
$ : int [1:4] 25 28 31 54
$ : int [1:4] 24 28 33 81
$ : int [1:4] 27 33 42 81
$ : int [1:2] 26 29
$ : int [1:6] 24 25 33 49 52 54
$ : int [1:2] 27 37
$ : int 33
$ : int [1:2] 24 36
$ : int 50
$ : int [1:5] 25 26 28 30 81
$ : int [1:3] 36 45 80
$ : int [1:6] 21 41 46 47 80 82
$ : int [1:5] 31 34 45 56 80
$ : int [1:2] 29 42
$ : int [1:3] 44 77 79
$ : int [1:4] 40 42 43 81
$ : int [1:3] 39 45 79
$ : int [1:5] 23 35 45 79 82
$ : int [1:5] 26 37 39 43 81
$ : int [1:3] 39 42 44
$ : int [1:2] 38 43
$ : int [1:6] 34 36 40 41 79 80
$ : int [1:5] 8 9 35 47 86
$ : int [1:5] 8 35 46 80 86
$ : int [1:5] 50 51 52 53 55
$ : int [1:4] 28 51 52 54
$ : int [1:6] 32 48 51 52 54 55
$ : int [1:4] 48 49 50 52
$ : int [1:6] 28 48 49 50 51 54
$ : int [1:2] 48 55
$ : int [1:5] 24 28 49 50 52
$ : int [1:4] 48 50 53 75
$ : int 36
$ : int [1:5] 1 2 3 58 64
$ : int [1:5] 2 57 64 66 68
$ : int [1:3] 60 87 88
$ : int [1:4] 12 13 59 61
$ : int [1:5] 12 60 62 63 87
$ : int [1:4] 61 63 77 87
$ : int [1:5] 12 18 61 62 83
$ : int [1:4] 1 57 58 76
$ : int 76
$ : int [1:5] 58 67 68 76 84
$ : int [1:2] 7 66
$ : int [1:4] 9 58 66 84
$ : int [1:2] 6 75
$ : int [1:3] 10 72 73
$ : int [1:2] 73 74
$ : int [1:3] 10 11 70
$ : int [1:4] 19 70 71 74
$ : int [1:5] 19 21 71 73 86
$ : int [1:2] 55 69
$ : int [1:3] 64 65 66
$ : int [1:3] 23 38 62
$ : int [1:2] 2 8
$ : int [1:4] 38 40 41 45
$ : int [1:5] 34 35 36 45 47
$ : int [1:5] 25 26 33 39 42
$ : int [1:6] 19 20 21 23 35 41
$ : int [1:4] 12 13 16 63
$ : int [1:4] 7 9 66 68
$ : int [1:2] 2 5
$ : int [1:4] 21 46 47 74
$ : int [1:4] 59 61 62 88
$ : int [1:2] 59 87
- attr(*, "class")= chr "nb"
- attr(*, "region.id")= chr [1:88] "1" "2" "3" "4" ...
- attr(*, "call")= language dnearneigh(x = coords, d1 = 0, d2 = 62, longlat = TRUE)
- attr(*, "dnn")= num [1:2] 0 62
- attr(*, "bounds")= chr [1:2] "GE" "LE"
- attr(*, "nbtype")= chr "distance"
- attr(*, "sym")= logi TRUEAnother way to display the structure of the weight matrix is to combine table() and card() from spdep package.
table(hunan$County, card(wm_d62))
1 2 3 4 5 6
Anhua 1 0 0 0 0 0
Anren 0 0 0 1 0 0
Anxiang 0 0 0 0 1 0
Baojing 0 0 0 0 1 0
Chaling 0 0 1 0 0 0
Changning 0 0 1 0 0 0
Changsha 0 0 0 1 0 0
Chengbu 0 1 0 0 0 0
Chenxi 0 0 0 1 0 0
Cili 0 1 0 0 0 0
Dao 0 0 0 1 0 0
Dongan 0 0 1 0 0 0
Dongkou 0 0 0 1 0 0
Fenghuang 0 0 0 1 0 0
Guidong 0 0 1 0 0 0
Guiyang 0 0 0 1 0 0
Guzhang 0 0 0 0 0 1
Hanshou 0 0 0 1 0 0
Hengdong 0 0 0 0 1 0
Hengnan 0 0 0 0 1 0
Hengshan 0 0 0 0 0 1
Hengyang 0 0 0 0 0 1
Hongjiang 0 0 0 0 1 0
Huarong 0 0 0 1 0 0
Huayuan 0 0 0 1 0 0
Huitong 0 0 0 1 0 0
Jiahe 0 0 0 0 1 0
Jianghua 0 0 1 0 0 0
Jiangyong 0 1 0 0 0 0
Jingzhou 0 1 0 0 0 0
Jinshi 0 0 0 1 0 0
Jishou 0 0 0 0 0 1
Lanshan 0 0 0 1 0 0
Leiyang 0 0 0 1 0 0
Lengshuijiang 0 0 1 0 0 0
Li 0 0 1 0 0 0
Lianyuan 0 0 0 0 1 0
Liling 0 1 0 0 0 0
Linli 0 0 0 1 0 0
Linwu 0 0 0 1 0 0
Linxiang 1 0 0 0 0 0
Liuyang 0 1 0 0 0 0
Longhui 0 0 1 0 0 0
Longshan 0 1 0 0 0 0
Luxi 0 0 0 0 1 0
Mayang 0 0 0 0 0 1
Miluo 0 0 0 0 1 0
Nan 0 0 0 0 1 0
Ningxiang 0 0 0 1 0 0
Ningyuan 0 0 0 0 1 0
Pingjiang 0 1 0 0 0 0
Qidong 0 0 1 0 0 0
Qiyang 0 0 1 0 0 0
Rucheng 0 1 0 0 0 0
Sangzhi 0 1 0 0 0 0
Shaodong 0 0 0 0 1 0
Shaoshan 0 0 0 0 1 0
Shaoyang 0 0 0 1 0 0
Shimen 1 0 0 0 0 0
Shuangfeng 0 0 0 0 0 1
Shuangpai 0 0 0 1 0 0
Suining 0 0 0 0 1 0
Taojiang 0 1 0 0 0 0
Taoyuan 0 1 0 0 0 0
Tongdao 0 1 0 0 0 0
Wangcheng 0 0 0 1 0 0
Wugang 0 0 1 0 0 0
Xiangtan 0 0 0 1 0 0
Xiangxiang 0 0 0 0 1 0
Xiangyin 0 0 0 1 0 0
Xinhua 0 0 0 0 1 0
Xinhuang 1 0 0 0 0 0
Xinning 0 1 0 0 0 0
Xinshao 0 0 0 0 0 1
Xintian 0 0 0 0 1 0
Xupu 0 1 0 0 0 0
Yanling 0 0 1 0 0 0
Yizhang 1 0 0 0 0 0
Yongshun 0 0 0 1 0 0
Yongxing 0 0 0 1 0 0
You 0 0 0 1 0 0
Yuanjiang 0 0 0 0 1 0
Yuanling 1 0 0 0 0 0
Yueyang 0 0 1 0 0 0
Zhijiang 0 0 0 0 1 0
Zhongfang 0 0 0 1 0 0
Zhuzhou 0 0 0 0 1 0
Zixing 0 0 1 0 0 0n_comp <- n.comp.nb(wm_d62)
n_comp$n[1] 1table(n_comp$comp.id)
1
88 6.2.1 Plotting fixed distance weight matrix
Next, we will plot the distance weight matrix by using the code chunk below.
plot(hunan$geometry, border="lightgrey")
plot(wm_d62, coords, add=TRUE)
plot(k1, coords, add=TRUE, col="red", length=0.08)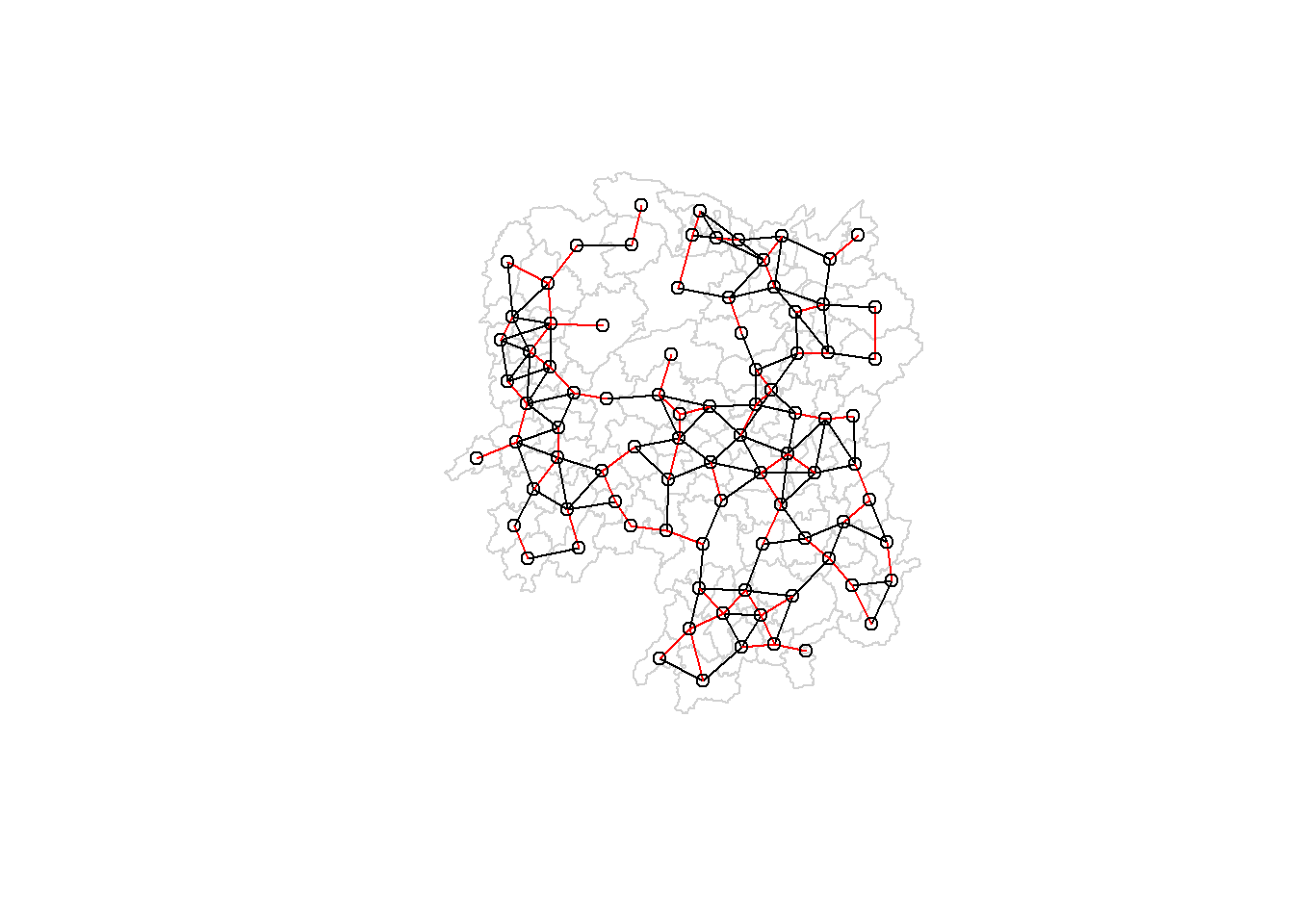
The red lines show the links of 1st nearest neighbours and the black lines show the links of neighbours within the cut-off distance of 62km.
Alternatively, we can plot both of them next to each other by using the code chunk below.
par(mfrow=c(1,2), mar = c(0, 0, 2, 3) + 0.1)
plot(hunan$geometry, border="lightgrey", main="1st nearest neighbours")
plot(k1, coords, add=TRUE, col="red", length=0.08, main="1st nearest neighbours")
title(main = '1st nearest neighbours')
plot(hunan$geometry, border="lightgrey")
plot(wm_d62, coords, add=TRUE, pch = 19, cex = 0.6)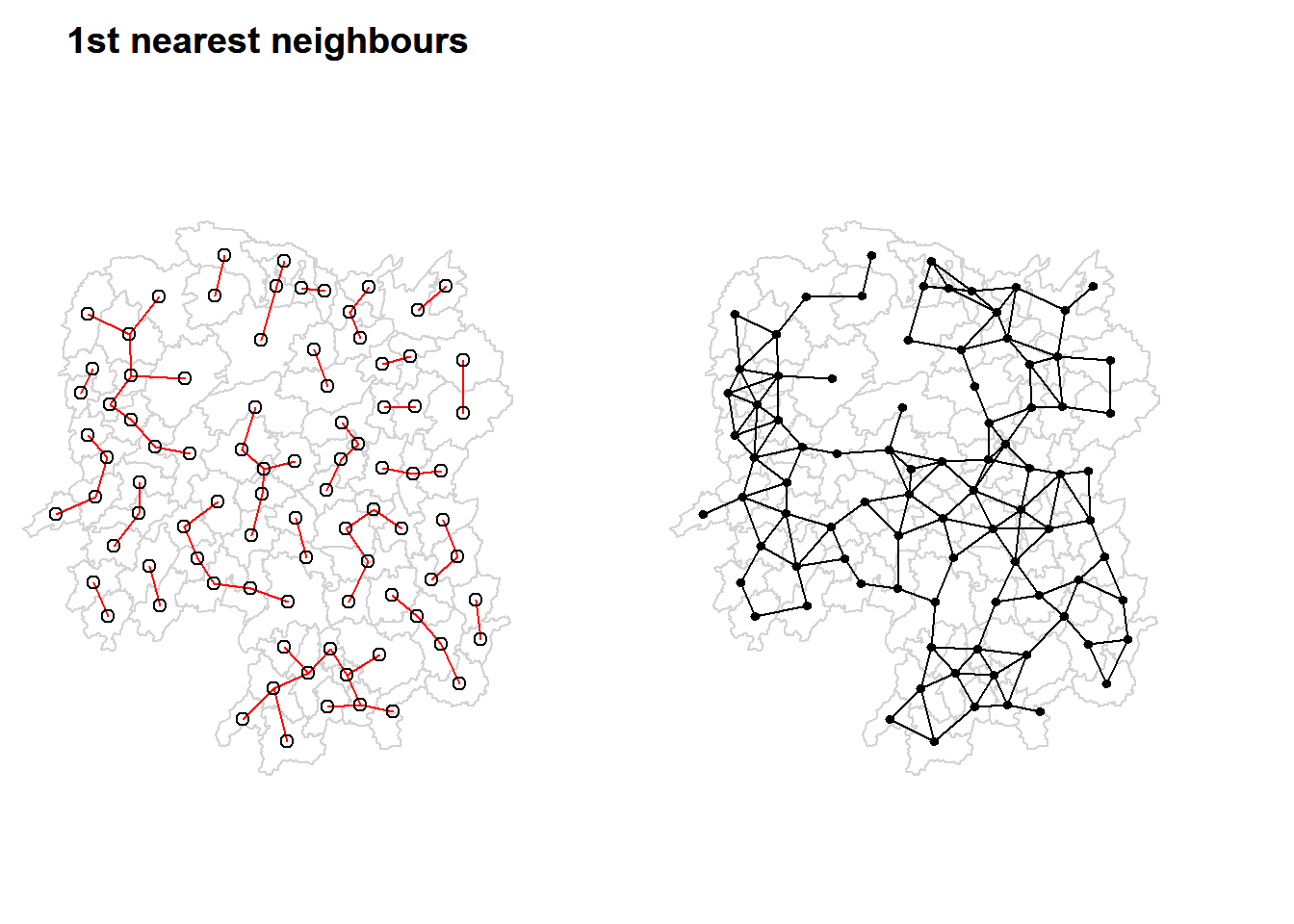
6.3 Computing adaptive distance weight matrix
One of the characteristics of fixed distance weight matrix is that more densely settled areas (usually the urban areas) tend to have more neighbours and the less densely settled areas (usually the rural counties) tend to have lesser neighbours. Having many neighbours smoothes the neighbour relationship across more neighbours.
It is possible to control the numbers of neighbours directly using k-nearest neighbours, either accepting asymmetric neighbours or imposing symmetry as shown in the code chunk below.
knn6 <- knn2nb(knearneigh(coords, k=6))
knn6Neighbour list object:
Number of regions: 88
Number of nonzero links: 528
Percentage nonzero weights: 6.818182
Average number of links: 6
Non-symmetric neighbours listSimilarly, we can display the content of the matrix by using str().
str(knn6)List of 88
$ : int [1:6] 2 3 4 5 57 64
$ : int [1:6] 1 3 57 58 78 85
$ : int [1:6] 1 2 4 5 57 85
$ : int [1:6] 1 3 5 6 69 85
$ : int [1:6] 1 3 4 6 69 85
$ : int [1:6] 3 4 5 69 75 85
$ : int [1:6] 9 66 67 71 74 84
$ : int [1:6] 9 46 47 78 80 86
$ : int [1:6] 8 46 66 68 84 86
$ : int [1:6] 16 19 22 70 72 73
$ : int [1:6] 10 14 16 17 70 72
$ : int [1:6] 13 15 60 61 63 83
$ : int [1:6] 12 15 60 61 63 83
$ : int [1:6] 11 15 16 17 72 83
$ : int [1:6] 12 13 14 17 60 83
$ : int [1:6] 10 11 17 22 72 83
$ : int [1:6] 10 11 14 16 72 83
$ : int [1:6] 20 22 23 63 77 83
$ : int [1:6] 10 20 21 73 74 82
$ : int [1:6] 18 19 21 22 23 82
$ : int [1:6] 19 20 35 74 82 86
$ : int [1:6] 10 16 18 19 20 83
$ : int [1:6] 18 20 41 77 79 82
$ : int [1:6] 25 28 31 52 54 81
$ : int [1:6] 24 28 31 33 54 81
$ : int [1:6] 25 27 29 33 42 81
$ : int [1:6] 26 29 30 37 42 81
$ : int [1:6] 24 25 33 49 52 54
$ : int [1:6] 26 27 37 42 43 81
$ : int [1:6] 26 27 28 33 49 81
$ : int [1:6] 24 25 36 39 40 54
$ : int [1:6] 24 31 50 54 55 56
$ : int [1:6] 25 26 28 30 49 81
$ : int [1:6] 36 40 41 45 56 80
$ : int [1:6] 21 41 46 47 80 82
$ : int [1:6] 31 34 40 45 56 80
$ : int [1:6] 26 27 29 42 43 44
$ : int [1:6] 23 43 44 62 77 79
$ : int [1:6] 25 40 42 43 44 81
$ : int [1:6] 31 36 39 43 45 79
$ : int [1:6] 23 35 45 79 80 82
$ : int [1:6] 26 27 37 39 43 81
$ : int [1:6] 37 39 40 42 44 79
$ : int [1:6] 37 38 39 42 43 79
$ : int [1:6] 34 36 40 41 79 80
$ : int [1:6] 8 9 35 47 78 86
$ : int [1:6] 8 21 35 46 80 86
$ : int [1:6] 49 50 51 52 53 55
$ : int [1:6] 28 33 48 51 52 54
$ : int [1:6] 32 48 51 52 54 55
$ : int [1:6] 28 48 49 50 52 54
$ : int [1:6] 28 48 49 50 51 54
$ : int [1:6] 48 50 51 52 55 75
$ : int [1:6] 24 28 49 50 51 52
$ : int [1:6] 32 48 50 52 53 75
$ : int [1:6] 32 34 36 78 80 85
$ : int [1:6] 1 2 3 58 64 68
$ : int [1:6] 2 57 64 66 68 78
$ : int [1:6] 12 13 60 61 87 88
$ : int [1:6] 12 13 59 61 63 87
$ : int [1:6] 12 13 60 62 63 87
$ : int [1:6] 12 38 61 63 77 87
$ : int [1:6] 12 18 60 61 62 83
$ : int [1:6] 1 3 57 58 68 76
$ : int [1:6] 58 64 66 67 68 76
$ : int [1:6] 9 58 67 68 76 84
$ : int [1:6] 7 65 66 68 76 84
$ : int [1:6] 9 57 58 66 78 84
$ : int [1:6] 4 5 6 32 75 85
$ : int [1:6] 10 16 19 22 72 73
$ : int [1:6] 7 19 73 74 84 86
$ : int [1:6] 10 11 14 16 17 70
$ : int [1:6] 10 19 21 70 71 74
$ : int [1:6] 19 21 71 73 84 86
$ : int [1:6] 6 32 50 53 55 69
$ : int [1:6] 58 64 65 66 67 68
$ : int [1:6] 18 23 38 61 62 63
$ : int [1:6] 2 8 9 46 58 68
$ : int [1:6] 38 40 41 43 44 45
$ : int [1:6] 34 35 36 41 45 47
$ : int [1:6] 25 26 28 33 39 42
$ : int [1:6] 19 20 21 23 35 41
$ : int [1:6] 12 13 15 16 22 63
$ : int [1:6] 7 9 66 68 71 74
$ : int [1:6] 2 3 4 5 56 69
$ : int [1:6] 8 9 21 46 47 74
$ : int [1:6] 59 60 61 62 63 88
$ : int [1:6] 59 60 61 62 63 87
- attr(*, "region.id")= chr [1:88] "1" "2" "3" "4" ...
- attr(*, "call")= language knearneigh(x = coords, k = 6)
- attr(*, "sym")= logi FALSE
- attr(*, "type")= chr "knn"
- attr(*, "knn-k")= num 6
- attr(*, "class")= chr "nb"In this way, each county has the same number of neighbours at exactly six neighbours!
6.3.1 Plotting distance based neighbours
We can plot the weight matrix using the code chunk below.
plot(hunan$geometry, border="lightgrey")
plot(knn6, coords, pch = 19, cex = 0.6, add = TRUE, col = "red")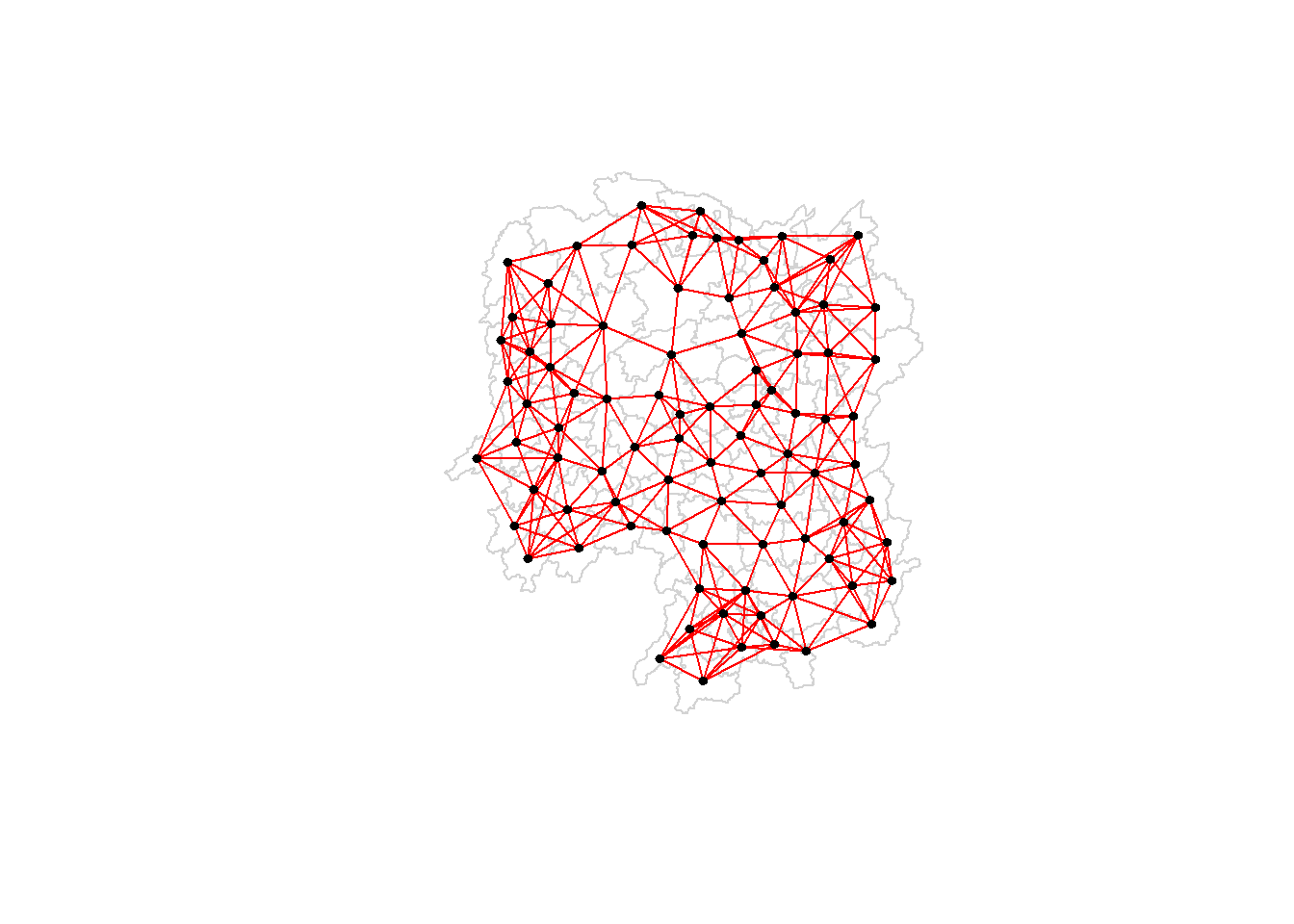
7 Weights based on IDW
In this section, we will derive a spatial weight matrix based on Inversed Distance method.
First, we will compute the distances between areas by using nbdists() from spdep package.
dist <- nbdists(wm_q, coords, longlat = TRUE)
ids <- lapply(dist, function(x) 1/(x))
ids[[1]]
[1] 0.01535405 0.03916350 0.01820896 0.02807922 0.01145113
[[2]]
[1] 0.01535405 0.01764308 0.01925924 0.02323898 0.01719350
[[3]]
[1] 0.03916350 0.02822040 0.03695795 0.01395765
[[4]]
[1] 0.01820896 0.02822040 0.03414741 0.01539065
[[5]]
[1] 0.03695795 0.03414741 0.01524598 0.01618354
[[6]]
[1] 0.015390649 0.015245977 0.021748129 0.011883901 0.009810297
[[7]]
[1] 0.01708612 0.01473997 0.01150924 0.01872915
[[8]]
[1] 0.02022144 0.03453056 0.02529256 0.01036340 0.02284457 0.01500600 0.01515314
[[9]]
[1] 0.02022144 0.01574888 0.02109502 0.01508028 0.02902705 0.01502980
[[10]]
[1] 0.02281552 0.01387777 0.01538326 0.01346650 0.02100510 0.02631658 0.01874863
[8] 0.01500046
[[11]]
[1] 0.01882869 0.02243492 0.02247473
[[12]]
[1] 0.02779227 0.02419652 0.02333385 0.02986130 0.02335429
[[13]]
[1] 0.02779227 0.02650020 0.02670323 0.01714243
[[14]]
[1] 0.01882869 0.01233868 0.02098555
[[15]]
[1] 0.02650020 0.01233868 0.01096284 0.01562226
[[16]]
[1] 0.02281552 0.02466962 0.02765018 0.01476814 0.01671430
[[17]]
[1] 0.01387777 0.02243492 0.02098555 0.01096284 0.02466962 0.01593341 0.01437996
[[18]]
[1] 0.02039779 0.02032767 0.01481665 0.01473691 0.01459380
[[19]]
[1] 0.01538326 0.01926323 0.02668415 0.02140253 0.01613589 0.01412874
[[20]]
[1] 0.01346650 0.02039779 0.01926323 0.01723025 0.02153130 0.01469240 0.02327034
[[21]]
[1] 0.02668415 0.01723025 0.01766299 0.02644986 0.02163800
[[22]]
[1] 0.02100510 0.02765018 0.02032767 0.02153130 0.01489296
[[23]]
[1] 0.01481665 0.01469240 0.01401432 0.02246233 0.01880425 0.01530458 0.01849605
[[24]]
[1] 0.02354598 0.01837201 0.02607264 0.01220154 0.02514180
[[25]]
[1] 0.02354598 0.02188032 0.01577283 0.01949232 0.02947957
[[26]]
[1] 0.02155798 0.01745522 0.02212108 0.02220532
[[27]]
[1] 0.02155798 0.02490625 0.01562326
[[28]]
[1] 0.01837201 0.02188032 0.02229549 0.03076171 0.02039506
[[29]]
[1] 0.02490625 0.01686587 0.01395022
[[30]]
[1] 0.02090587
[[31]]
[1] 0.02607264 0.01577283 0.01219005 0.01724850 0.01229012 0.01609781 0.01139438
[8] 0.01150130
[[32]]
[1] 0.01220154 0.01219005 0.01712515 0.01340413 0.01280928 0.01198216 0.01053374
[8] 0.01065655
[[33]]
[1] 0.01949232 0.01745522 0.02229549 0.02090587 0.01979045
[[34]]
[1] 0.03113041 0.03589551 0.02882915
[[35]]
[1] 0.01766299 0.02185795 0.02616766 0.02111721 0.02108253 0.01509020
[[36]]
[1] 0.01724850 0.03113041 0.01571707 0.01860991 0.02073549 0.01680129
[[37]]
[1] 0.01686587 0.02234793 0.01510990 0.01550676
[[38]]
[1] 0.01401432 0.02407426 0.02276151 0.01719415
[[39]]
[1] 0.01229012 0.02172543 0.01711924 0.02629732 0.01896385
[[40]]
[1] 0.01609781 0.01571707 0.02172543 0.01506473 0.01987922 0.01894207
[[41]]
[1] 0.02246233 0.02185795 0.02205991 0.01912542 0.01601083 0.01742892
[[42]]
[1] 0.02212108 0.01562326 0.01395022 0.02234793 0.01711924 0.01836831 0.01683518
[[43]]
[1] 0.01510990 0.02629732 0.01506473 0.01836831 0.03112027 0.01530782
[[44]]
[1] 0.01550676 0.02407426 0.03112027 0.01486508
[[45]]
[1] 0.03589551 0.01860991 0.01987922 0.02205991 0.02107101 0.01982700
[[46]]
[1] 0.03453056 0.04033752 0.02689769
[[47]]
[1] 0.02529256 0.02616766 0.04033752 0.01949145 0.02181458
[[48]]
[1] 0.02313819 0.03370576 0.02289485 0.01630057 0.01818085
[[49]]
[1] 0.03076171 0.02138091 0.02394529 0.01990000
[[50]]
[1] 0.01712515 0.02313819 0.02551427 0.02051530 0.02187179
[[51]]
[1] 0.03370576 0.02138091 0.02873854
[[52]]
[1] 0.02289485 0.02394529 0.02551427 0.02873854 0.03516672
[[53]]
[1] 0.01630057 0.01979945 0.01253977
[[54]]
[1] 0.02514180 0.02039506 0.01340413 0.01990000 0.02051530 0.03516672
[[55]]
[1] 0.01280928 0.01818085 0.02187179 0.01979945 0.01882298
[[56]]
[1] 0.01036340 0.01139438 0.01198216 0.02073549 0.01214479 0.01362855 0.01341697
[[57]]
[1] 0.028079221 0.017643082 0.031423501 0.029114131 0.013520292 0.009903702
[[58]]
[1] 0.01925924 0.03142350 0.02722997 0.01434859 0.01567192
[[59]]
[1] 0.01696711 0.01265572 0.01667105 0.01785036
[[60]]
[1] 0.02419652 0.02670323 0.01696711 0.02343040
[[61]]
[1] 0.02333385 0.01265572 0.02343040 0.02514093 0.02790764 0.01219751 0.02362452
[[62]]
[1] 0.02514093 0.02002219 0.02110260
[[63]]
[1] 0.02986130 0.02790764 0.01407043 0.01805987
[[64]]
[1] 0.02911413 0.01689892
[[65]]
[1] 0.02471705
[[66]]
[1] 0.01574888 0.01726461 0.03068853 0.01954805 0.01810569
[[67]]
[1] 0.01708612 0.01726461 0.01349843 0.01361172
[[68]]
[1] 0.02109502 0.02722997 0.03068853 0.01406357 0.01546511
[[69]]
[1] 0.02174813 0.01645838 0.01419926
[[70]]
[1] 0.02631658 0.01963168 0.02278487
[[71]]
[1] 0.01473997 0.01838483 0.03197403
[[72]]
[1] 0.01874863 0.02247473 0.01476814 0.01593341 0.01963168
[[73]]
[1] 0.01500046 0.02140253 0.02278487 0.01838483 0.01652709
[[74]]
[1] 0.01150924 0.01613589 0.03197403 0.01652709 0.01342099 0.02864567
[[75]]
[1] 0.011883901 0.010533736 0.012539774 0.018822977 0.016458383 0.008217581
[[76]]
[1] 0.01352029 0.01434859 0.01689892 0.02471705 0.01954805 0.01349843 0.01406357
[[77]]
[1] 0.014736909 0.018804247 0.022761507 0.012197506 0.020022195 0.014070428
[7] 0.008440896
[[78]]
[1] 0.02323898 0.02284457 0.01508028 0.01214479 0.01567192 0.01546511 0.01140779
[[79]]
[1] 0.01530458 0.01719415 0.01894207 0.01912542 0.01530782 0.01486508 0.02107101
[[80]]
[1] 0.01500600 0.02882915 0.02111721 0.01680129 0.01601083 0.01982700 0.01949145
[8] 0.01362855
[[81]]
[1] 0.02947957 0.02220532 0.01150130 0.01979045 0.01896385 0.01683518
[[82]]
[1] 0.02327034 0.02644986 0.01849605 0.02108253 0.01742892
[[83]]
[1] 0.023354289 0.017142433 0.015622258 0.016714303 0.014379961 0.014593799
[7] 0.014892965 0.018059871 0.008440896
[[84]]
[1] 0.01872915 0.02902705 0.01810569 0.01361172 0.01342099 0.01297994
[[85]]
[1] 0.011451133 0.017193502 0.013957649 0.016183544 0.009810297 0.010656545
[7] 0.013416965 0.009903702 0.014199260 0.008217581 0.011407794
[[86]]
[1] 0.01515314 0.01502980 0.01412874 0.02163800 0.01509020 0.02689769 0.02181458
[8] 0.02864567 0.01297994
[[87]]
[1] 0.01667105 0.02362452 0.02110260 0.02058034
[[88]]
[1] 0.01785036 0.020580347.1 Spatial lag with row-standardized weights
Next, we need to assign weights to each neighboring polygon. In our case, each neighboring polygon will be assigned equal weight (using nb2listw() argument style=“W”). This is accomplished by assigning the fraction 1/(number of neighbours) to each neighboring county then summing the weighted income values. While this is the most intuitive way to summarise the neighbors’ values. One drawback of this approach is that polygons along the edges of the study area will base their lagged values on fewer polygons and this can potentially over- or under-estimate the true spatial autocorrelation in the data. For simplicity, we will use the style=“W” option for our example. There are other more robust options available, notably style=“B”.
rswm_q <- nb2listw(wm_q, style="W", zero.policy = TRUE)
rswm_qCharacteristics of weights list object:
Neighbour list object:
Number of regions: 88
Number of nonzero links: 448
Percentage nonzero weights: 5.785124
Average number of links: 5.090909
Weights style: W
Weights constants summary:
n nn S0 S1 S2
W 88 7744 88 37.86334 365.9147The zero.policy=TRUE option allows for lists of non-neighbors. This should be used with caution since the user may not be aware of missing neighbors in their dataset however, a zero.policy of FALSE would return an error.
To see the weight of the first polygon’s neighbors type:
rswm_q$weights[1][[1]]
[1] 0.2 0.2 0.2 0.2 0.2Each neighbor is assigned a 0.2 of the total weight. This means that when R computes the average neighboring income values, each neighbor’s income will be multiplied by 0.2 before being tallied.
Using the same method, we can also derive a row standardised distance weight matrix by using the code chunk below.
rswm_ids <- nb2listw(wm_q, glist=ids, style="B", zero.policy=TRUE)
rswm_idsCharacteristics of weights list object:
Neighbour list object:
Number of regions: 88
Number of nonzero links: 448
Percentage nonzero weights: 5.785124
Average number of links: 5.090909
Weights style: B
Weights constants summary:
n nn S0 S1 S2
B 88 7744 8.786867 0.3776535 3.8137rswm_ids$weights[1][[1]]
[1] 0.01535405 0.03916350 0.01820896 0.02807922 0.01145113summary(unlist(rswm_ids$weights)) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.008218 0.015088 0.018739 0.019614 0.022823 0.040338 8 Application of Spatial Weight Matrix
In this section, we will demonstrate how to create three different spatial lagged variables, they are:
spatial lag with row-standardized weights,
spatial lag as a sum of neighbouring values,
spatial window average, and spatial window sum.
8.1 Spatial lag with row-standardized weights
Finally, we’ll compute the average neighbor GDPPC value for each polygon. These values are often referred to as spatially lagged values.
GDPPC.lag <- lag.listw(rswm_q, hunan$GDPPC)
GDPPC.lag [1] 24847.20 22724.80 24143.25 27737.50 27270.25 21248.80 43747.00 33582.71
[9] 45651.17 32027.62 32671.00 20810.00 25711.50 30672.33 33457.75 31689.20
[17] 20269.00 23901.60 25126.17 21903.43 22718.60 25918.80 20307.00 20023.80
[25] 16576.80 18667.00 14394.67 19848.80 15516.33 20518.00 17572.00 15200.12
[33] 18413.80 14419.33 24094.50 22019.83 12923.50 14756.00 13869.80 12296.67
[41] 15775.17 14382.86 11566.33 13199.50 23412.00 39541.00 36186.60 16559.60
[49] 20772.50 19471.20 19827.33 15466.80 12925.67 18577.17 14943.00 24913.00
[57] 25093.00 24428.80 17003.00 21143.75 20435.00 17131.33 24569.75 23835.50
[65] 26360.00 47383.40 55157.75 37058.00 21546.67 23348.67 42323.67 28938.60
[73] 25880.80 47345.67 18711.33 29087.29 20748.29 35933.71 15439.71 29787.50
[81] 18145.00 21617.00 29203.89 41363.67 22259.09 44939.56 16902.00 16930.00With reference to the following code chunk where we retrieve the GDPPC of the five neighbouring counties of the first county in our data, Anxiang:
nb1 <- wm_q[[1]]
nb1 <- hunan$GDPPC[nb1]
nb1[1] 20981 34592 24473 21311 22879sum(nb1) / 5[1] 24847.2We can see that Spatial lag with row-standardized weights gives each neighbour equal weight.
lag.list <- list(hunan$NAME_3, lag.listw(rswm_q, hunan$GDPPC))
lag.res <- as.data.frame(lag.list)
colnames(lag.res) <- c("NAME_3", "lag GDPPC")
hunan <- left_join(hunan,lag.res)Joining, by = "NAME_3"The following table shows the average neighboring income values (stored in the Inc.lag object) for each county.
head(hunan)Simple feature collection with 6 features and 36 fields
Geometry type: POLYGON
Dimension: XY
Bounding box: xmin: 110.4922 ymin: 28.61762 xmax: 112.3013 ymax: 30.12812
Geodetic CRS: WGS 84
NAME_2 ID_3 NAME_3 ENGTYPE_3 Shape_Leng Shape_Area County City
1 Changde 21098 Anxiang County 1.869074 0.10056190 Anxiang Changde
2 Changde 21100 Hanshou County 2.360691 0.19978745 Hanshou Changde
3 Changde 21101 Jinshi County City 1.425620 0.05302413 Jinshi Changde
4 Changde 21102 Li County 3.474325 0.18908121 Li Changde
5 Changde 21103 Linli County 2.289506 0.11450357 Linli Changde
6 Changde 21104 Shimen County 4.171918 0.37194707 Shimen Changde
avg_wage deposite FAI Gov_Rev Gov_Exp GDP GDPPC GIO Loan NIPCR
1 31935 5517.2 3541.0 243.64 1779.5 12482.0 23667 5108.9 2806.9 7693.7
2 32265 7979.0 8665.0 386.13 2062.4 15788.0 20981 13491.0 4550.0 8269.9
3 28692 4581.7 4777.0 373.31 1148.4 8706.9 34592 10935.0 2242.0 8169.9
4 32541 13487.0 16066.0 709.61 2459.5 20322.0 24473 18402.0 6748.0 8377.0
5 32667 564.1 7781.2 336.86 1538.7 10355.0 25554 8214.0 358.0 8143.1
6 33261 8334.4 10531.0 548.33 2178.8 16293.0 27137 17795.0 6026.5 6156.0
Bed Emp EmpR EmpRT Pri_Stu Sec_Stu Household Household_R NOIP Pop_R
1 1931 336.39 270.5 205.9 19.584 17.819 148.1 135.4 53 346.0
2 2560 456.78 388.8 246.7 42.097 33.029 240.2 208.7 95 553.2
3 848 122.78 82.1 61.7 8.723 7.592 81.9 43.7 77 92.4
4 2038 513.44 426.8 227.1 38.975 33.938 268.5 256.0 96 539.7
5 1440 307.36 272.2 100.8 23.286 18.943 129.1 157.2 99 246.6
6 2502 392.05 329.6 193.8 29.245 26.104 190.6 184.7 122 399.2
RSCG Pop_T Agri Service Disp_Inc RORP ROREmp lag GDPPC
1 3957.9 528.3 4524.41 14100 16610 0.6549309 0.8041262 24847.20
2 4460.5 804.6 6545.35 17727 18925 0.6875466 0.8511756 22724.80
3 3683.0 251.8 2562.46 7525 19498 0.3669579 0.6686757 24143.25
4 7110.2 832.5 7562.34 53160 18985 0.6482883 0.8312558 27737.50
5 3604.9 409.3 3583.91 7031 18604 0.6024921 0.8856065 27270.25
6 6490.7 600.5 5266.51 6981 19275 0.6647794 0.8407091 21248.80
geometry
1 POLYGON ((112.0625 29.75523...
2 POLYGON ((112.2288 29.11684...
3 POLYGON ((111.8927 29.6013,...
4 POLYGON ((111.3731 29.94649...
5 POLYGON ((111.6324 29.76288...
6 POLYGON ((110.8825 30.11675...Next, we will plot both the GDPPC and spatial lag GDPPC for comparison using the code chunk below.
gdppc <- qtm(hunan, "GDPPC")
lag_gdppc <- qtm(hunan, "lag GDPPC")
tmap_arrange(gdppc, lag_gdppc, asp=1, ncol=2)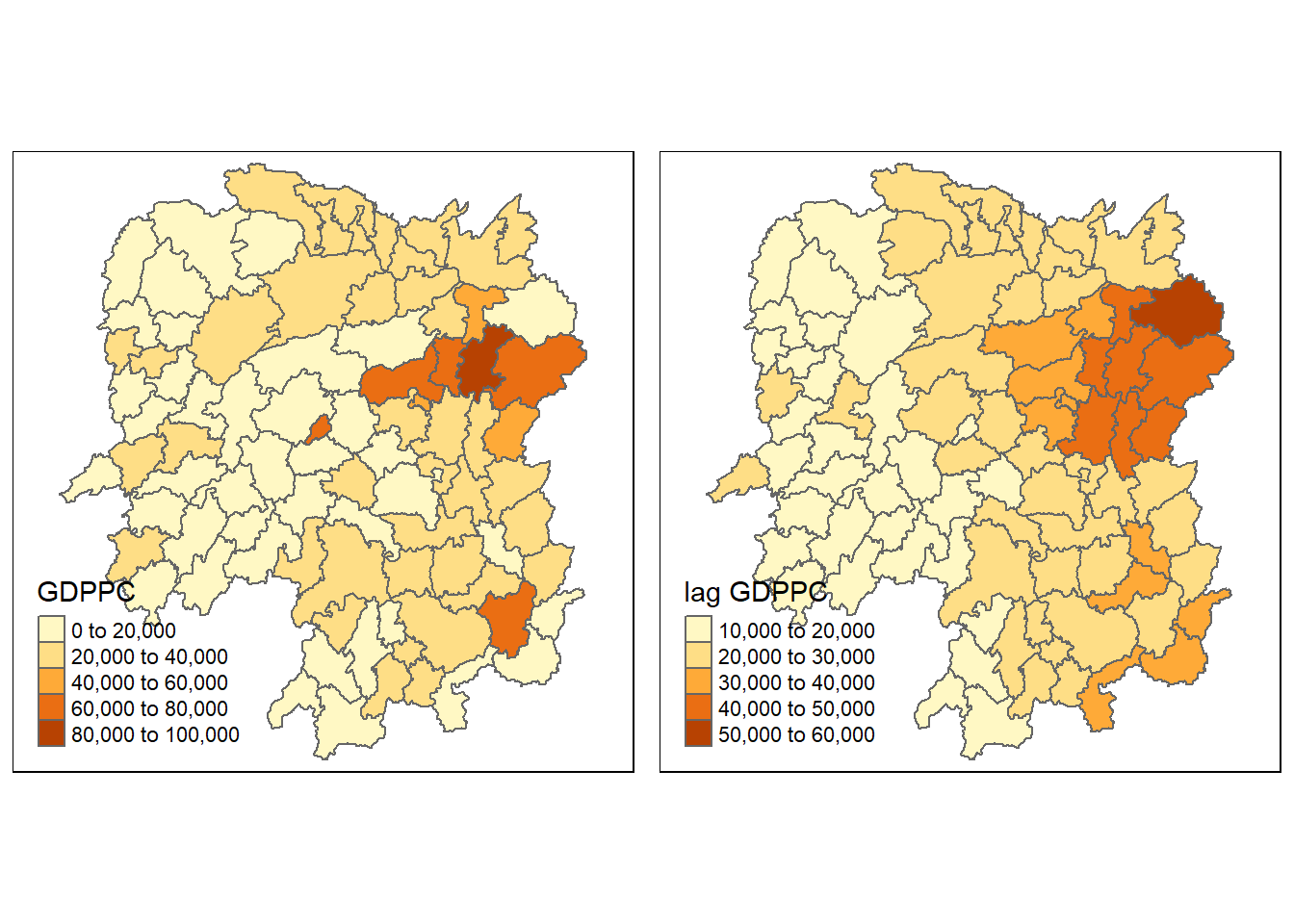
8.2 Spatial lag as a sum of neighboring values
We can calculate spatial lag as a sum of neighboring values by assigning binary weights. This requires us to go back to our neighbors list, then apply a function that will assign binary weights, then we use glist = in the nb2listw function to explicitly assign these weights.
We start by applying a function that will assign a value of 1 per each neighbor. This is done with lapply().
b_weights <- lapply(wm_q, function(x) 0*x + 1)
b_weights2 <- nb2listw(wm_q,
glist = b_weights,
style = "B")
b_weights2Characteristics of weights list object:
Neighbour list object:
Number of regions: 88
Number of nonzero links: 448
Percentage nonzero weights: 5.785124
Average number of links: 5.090909
Weights style: B
Weights constants summary:
n nn S0 S1 S2
B 88 7744 448 896 10224With the proper weights assigned, we can use lag.listw to compute a lag variable from our weight and GDPPC.
lag_sum <- list(hunan$NAME_3, lag.listw(b_weights2, hunan$GDPPC))
lag.res <- as.data.frame(lag_sum)
colnames(lag.res) <- c("NAME_3", "lag_sum GDPPC")First, let us examine the result by using the code chunk below.
lag_sum[[1]]
[1] "Anxiang" "Hanshou" "Jinshi" "Li"
[5] "Linli" "Shimen" "Liuyang" "Ningxiang"
[9] "Wangcheng" "Anren" "Guidong" "Jiahe"
[13] "Linwu" "Rucheng" "Yizhang" "Yongxing"
[17] "Zixing" "Changning" "Hengdong" "Hengnan"
[21] "Hengshan" "Leiyang" "Qidong" "Chenxi"
[25] "Zhongfang" "Huitong" "Jingzhou" "Mayang"
[29] "Tongdao" "Xinhuang" "Xupu" "Yuanling"
[33] "Zhijiang" "Lengshuijiang" "Shuangfeng" "Xinhua"
[37] "Chengbu" "Dongan" "Dongkou" "Longhui"
[41] "Shaodong" "Suining" "Wugang" "Xinning"
[45] "Xinshao" "Shaoshan" "Xiangxiang" "Baojing"
[49] "Fenghuang" "Guzhang" "Huayuan" "Jishou"
[53] "Longshan" "Luxi" "Yongshun" "Anhua"
[57] "Nan" "Yuanjiang" "Jianghua" "Lanshan"
[61] "Ningyuan" "Shuangpai" "Xintian" "Huarong"
[65] "Linxiang" "Miluo" "Pingjiang" "Xiangyin"
[69] "Cili" "Chaling" "Liling" "Yanling"
[73] "You" "Zhuzhou" "Sangzhi" "Yueyang"
[77] "Qiyang" "Taojiang" "Shaoyang" "Lianyuan"
[81] "Hongjiang" "Hengyang" "Guiyang" "Changsha"
[85] "Taoyuan" "Xiangtan" "Dao" "Jiangyong"
[[2]]
[1] 124236 113624 96573 110950 109081 106244 174988 235079 273907 256221
[11] 98013 104050 102846 92017 133831 158446 141883 119508 150757 153324
[21] 113593 129594 142149 100119 82884 74668 43184 99244 46549 20518
[31] 140576 121601 92069 43258 144567 132119 51694 59024 69349 73780
[41] 94651 100680 69398 52798 140472 118623 180933 82798 83090 97356
[51] 59482 77334 38777 111463 74715 174391 150558 122144 68012 84575
[61] 143045 51394 98279 47671 26360 236917 220631 185290 64640 70046
[71] 126971 144693 129404 284074 112268 203611 145238 251536 108078 238300
[81] 108870 108085 262835 248182 244850 404456 67608 33860nb1 <- wm_q[[1]]
nb1 <- hunan$GDPPC[nb1]
sum(nb1)[1] 124236We can see that Spatial lag as a sum of neighboring values simply sums the GDPPC values of all its neighbours.
Next, we will append the lag_sum GDPPC field into hunan sf data frame by using the code chunk below.
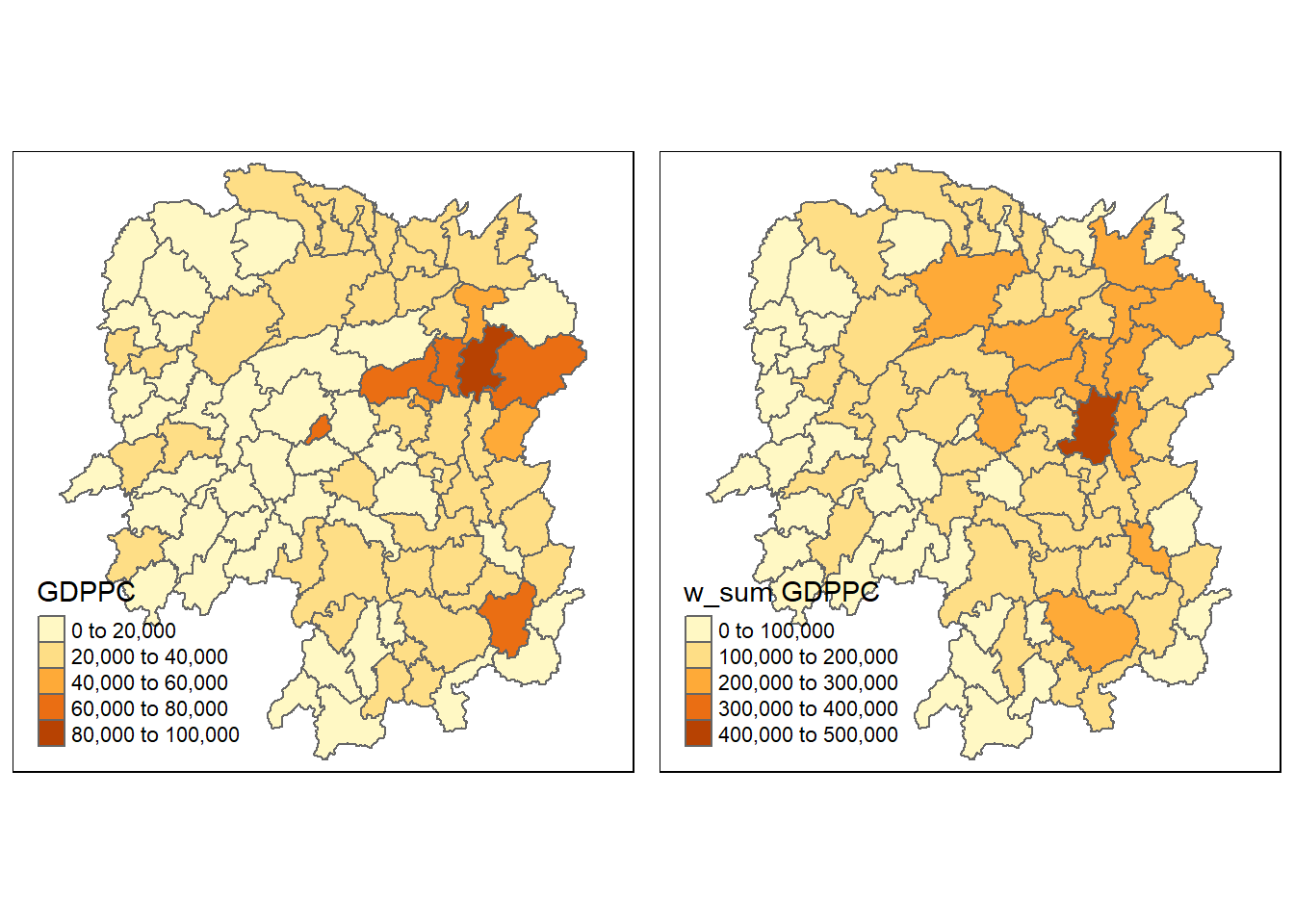
hunan <- left_join(hunan, lag.res)Joining, by = "NAME_3"Now, we can plot both the GDPPC and Spatial Lag Sum GDPPC for comparison using the code chunk below.
gdppc <- qtm(hunan, "GDPPC")
lag_sum_gdppc <- qtm(hunan, "lag_sum GDPPC")
tmap_arrange(gdppc, lag_sum_gdppc, asp=1, ncol=2)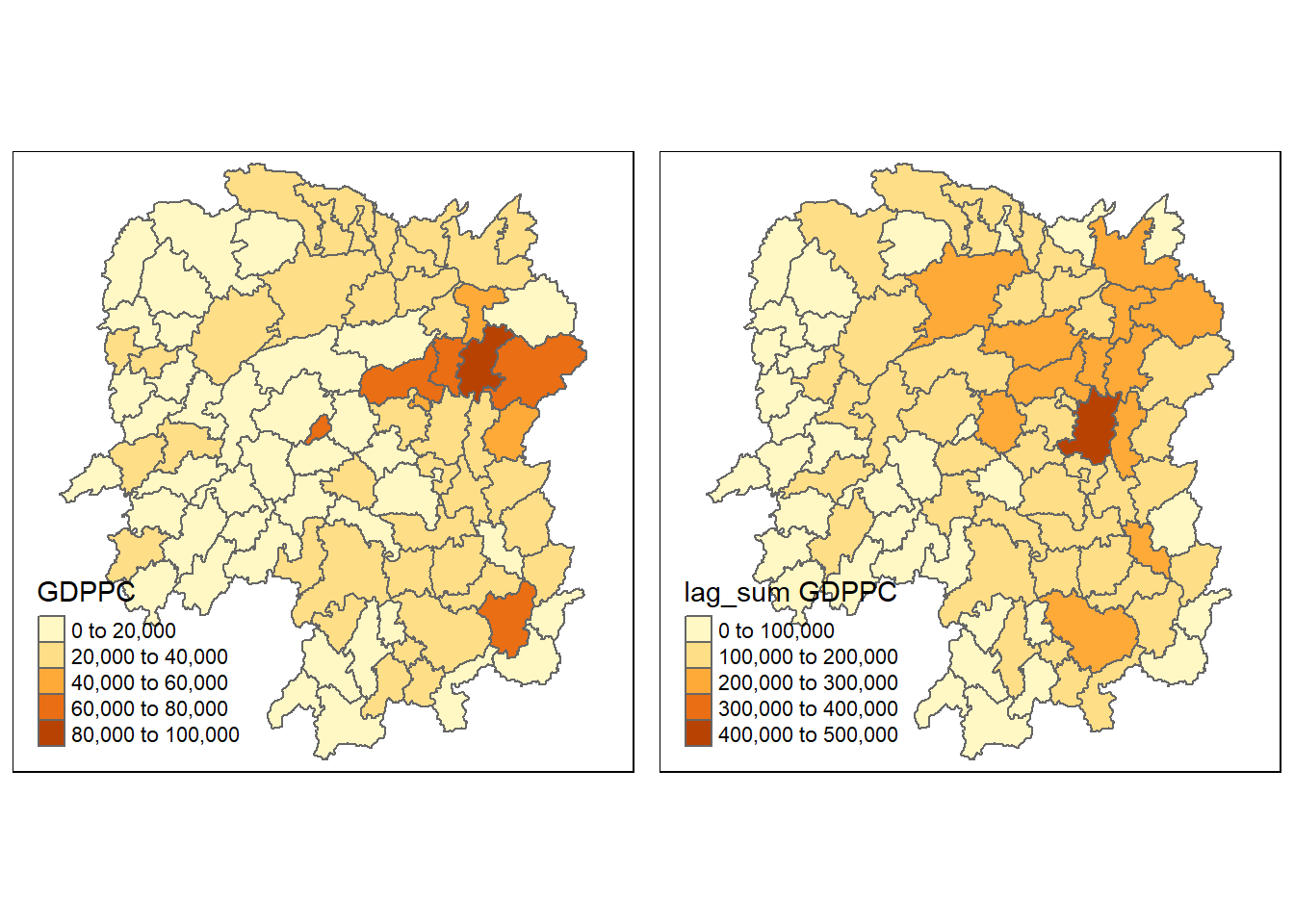
8.3 Spatial window average
The spatial window average uses row-standardized weights and includes the diagonal element - this means besides taking into consideration of its neighbours, this method also considers the county itself. To do this in R, we need to go back to the neighbors structure and add the diagonal element before assigning weights. To begin we assign this to a new variable because we will directly alter its structure to add the diagonal elements.
wm_q1 <- wm_qTo add the diagonal element to the neighbour list, we can use include.self() from spdep.
include.self(wm_q1)Neighbour list object:
Number of regions: 88
Number of nonzero links: 536
Percentage nonzero weights: 6.921488
Average number of links: 6.090909 Now we obtain weights using nb2listw()
wm_q1 <- nb2listw(wm_q1)
wm_q1Characteristics of weights list object:
Neighbour list object:
Number of regions: 88
Number of nonzero links: 448
Percentage nonzero weights: 5.785124
Average number of links: 5.090909
Weights style: W
Weights constants summary:
n nn S0 S1 S2
W 88 7744 88 37.86334 365.9147Lastly, we need to create the lag variable from our weight structure and GDPPC variable.
lag_w_avg_gpdpc <- lag.listw(wm_q1,
hunan$GDPPC)
lag_w_avg_gpdpc [1] 24847.20 22724.80 24143.25 27737.50 27270.25 21248.80 43747.00 33582.71
[9] 45651.17 32027.62 32671.00 20810.00 25711.50 30672.33 33457.75 31689.20
[17] 20269.00 23901.60 25126.17 21903.43 22718.60 25918.80 20307.00 20023.80
[25] 16576.80 18667.00 14394.67 19848.80 15516.33 20518.00 17572.00 15200.12
[33] 18413.80 14419.33 24094.50 22019.83 12923.50 14756.00 13869.80 12296.67
[41] 15775.17 14382.86 11566.33 13199.50 23412.00 39541.00 36186.60 16559.60
[49] 20772.50 19471.20 19827.33 15466.80 12925.67 18577.17 14943.00 24913.00
[57] 25093.00 24428.80 17003.00 21143.75 20435.00 17131.33 24569.75 23835.50
[65] 26360.00 47383.40 55157.75 37058.00 21546.67 23348.67 42323.67 28938.60
[73] 25880.80 47345.67 18711.33 29087.29 20748.29 35933.71 15439.71 29787.50
[81] 18145.00 21617.00 29203.89 41363.67 22259.09 44939.56 16902.00 16930.00Next, we will convert the lag variable listw object into a data.frame by using as.data.frame().
lag.list.wm_q1 <- list(hunan$NAME_3, lag.listw(wm_q1, hunan$GDPPC))
lag_wm_q1.res <- as.data.frame(lag.list.wm_q1)
colnames(lag_wm_q1.res) <- c("NAME_3", "lag_window_avg GDPPC")The third command line on the code chunk above renames the field names of lag_wm_q1.res object into NAME_3 and lag_window_avg GDPPC respectively.
Next, the code chunk below will be used to append lag_window_avg GDPPC values onto hunan sf data.frame by using left_join() of dplyr package.
hunan <- left_join(hunan, lag_wm_q1.res)Joining, by = "NAME_3"Lastly, qtm() from tmap package is used to plot the GDPPC and lag_window_avg GDPPC map next to each other for quick comparison.
gdppc <- qtm(hunan, "GDPPC")
w_avg_gdppc <- qtm(hunan, "lag_window_avg GDPPC")
tmap_arrange(gdppc, w_avg_gdppc, asp=1, ncol=2)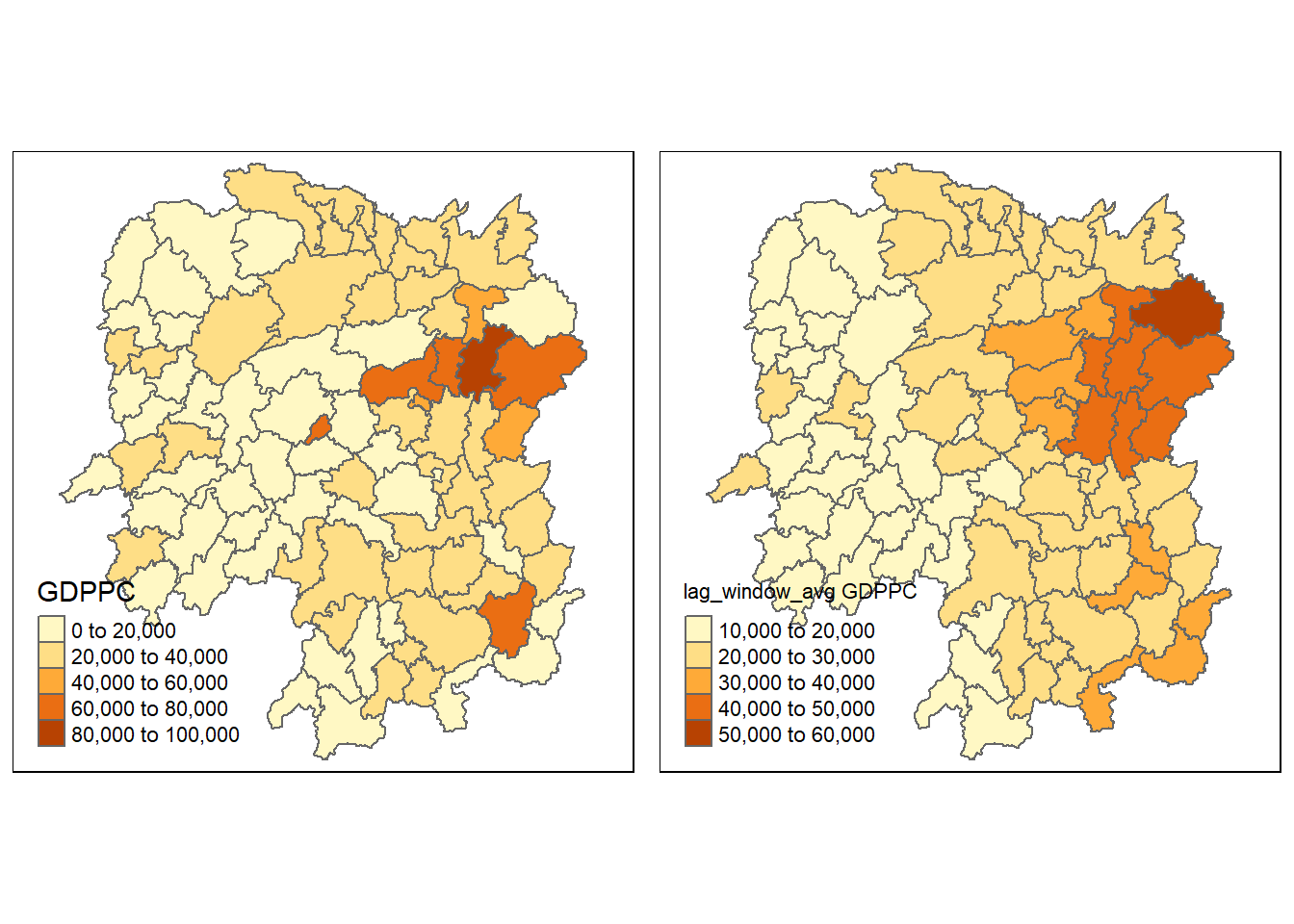
8.4 Spatial window sum
The spatial window sum is the counter part of the window average, but without using row-standardized weights. To do this we assign binary weights to the neighbor structure that includes the diagonal element.
wm_q1 <- wm_qTo add the diagonal element to the neighbour list, we use include.self() from spdep.
include.self(wm_q1)Neighbour list object:
Number of regions: 88
Number of nonzero links: 536
Percentage nonzero weights: 6.921488
Average number of links: 6.090909 wm_q1Neighbour list object:
Number of regions: 88
Number of nonzero links: 448
Percentage nonzero weights: 5.785124
Average number of links: 5.090909 Next, we will assign binary weights to the neighbour structure that includes the diagonal element.
b_weights <- lapply(wm_q1, function(x) 0*x + 1)
b_weights[1][[1]]
[1] 1 1 1 1 1Again, we use nb2listw() and glist() to explicitly assign weight values.
b_weights2 <- nb2listw(wm_q1,
glist = b_weights,
style = "B")
b_weights2Characteristics of weights list object:
Neighbour list object:
Number of regions: 88
Number of nonzero links: 448
Percentage nonzero weights: 5.785124
Average number of links: 5.090909
Weights style: B
Weights constants summary:
n nn S0 S1 S2
B 88 7744 448 896 10224With our new weight structure, we can compute the lag variable with lag.listw().
w_sum_gdppc <- list(hunan$NAME_3, lag.listw(b_weights2, hunan$GDPPC))
w_sum_gdppc[[1]]
[1] "Anxiang" "Hanshou" "Jinshi" "Li"
[5] "Linli" "Shimen" "Liuyang" "Ningxiang"
[9] "Wangcheng" "Anren" "Guidong" "Jiahe"
[13] "Linwu" "Rucheng" "Yizhang" "Yongxing"
[17] "Zixing" "Changning" "Hengdong" "Hengnan"
[21] "Hengshan" "Leiyang" "Qidong" "Chenxi"
[25] "Zhongfang" "Huitong" "Jingzhou" "Mayang"
[29] "Tongdao" "Xinhuang" "Xupu" "Yuanling"
[33] "Zhijiang" "Lengshuijiang" "Shuangfeng" "Xinhua"
[37] "Chengbu" "Dongan" "Dongkou" "Longhui"
[41] "Shaodong" "Suining" "Wugang" "Xinning"
[45] "Xinshao" "Shaoshan" "Xiangxiang" "Baojing"
[49] "Fenghuang" "Guzhang" "Huayuan" "Jishou"
[53] "Longshan" "Luxi" "Yongshun" "Anhua"
[57] "Nan" "Yuanjiang" "Jianghua" "Lanshan"
[61] "Ningyuan" "Shuangpai" "Xintian" "Huarong"
[65] "Linxiang" "Miluo" "Pingjiang" "Xiangyin"
[69] "Cili" "Chaling" "Liling" "Yanling"
[73] "You" "Zhuzhou" "Sangzhi" "Yueyang"
[77] "Qiyang" "Taojiang" "Shaoyang" "Lianyuan"
[81] "Hongjiang" "Hengyang" "Guiyang" "Changsha"
[85] "Taoyuan" "Xiangtan" "Dao" "Jiangyong"
[[2]]
[1] 124236 113624 96573 110950 109081 106244 174988 235079 273907 256221
[11] 98013 104050 102846 92017 133831 158446 141883 119508 150757 153324
[21] 113593 129594 142149 100119 82884 74668 43184 99244 46549 20518
[31] 140576 121601 92069 43258 144567 132119 51694 59024 69349 73780
[41] 94651 100680 69398 52798 140472 118623 180933 82798 83090 97356
[51] 59482 77334 38777 111463 74715 174391 150558 122144 68012 84575
[61] 143045 51394 98279 47671 26360 236917 220631 185290 64640 70046
[71] 126971 144693 129404 284074 112268 203611 145238 251536 108078 238300
[81] 108870 108085 262835 248182 244850 404456 67608 33860w_sum_gdppc.res <- as.data.frame(w_sum_gdppc)
colnames(w_sum_gdppc.res) <- c("NAME_3", "w_sum GDPPC")The second command line on the code chunk above renames the field names of w_sum_gdppc.res object into NAME_3 and w_sum GDPPC respectively.
Next, the code chunk below will be used to append w_sum GDPPC values onto hunan sf data.frame by using left_join() of dplyr package.
hunan <- left_join(hunan, w_sum_gdppc.res)Joining, by = "NAME_3"Lastly, qtm() of tmap package is used to plot the GDPPC and lag_sum GDPPC map next to each other for quick comparison.
gdppc <- qtm(hunan, "GDPPC")
w_sum_gdppc <- qtm(hunan, "w_sum GDPPC")
tmap_arrange(gdppc, w_sum_gdppc, asp=1, ncol=2)